Table of Contents
Plotting grain orientations
Typically, grain orientations are plotted with MTeX based on the Euler angles of each grain.
Extracting Euler angles
Extracting a list of Euler angles for the indexed grains can be done using timelessExtractEulerAngles, which is part of the TIMEleSS tools.
You will have a list of Bunge Euler angles (using a Bunge convention) as below. You can simply save it in a text file.
-31.88 273.49 206.05 -55.09 133.91 157.65 83.39 235.30 204.46 -47.58 78.99 149.71 -60.87 17.28 33.07
Loading and plotting grain orientations in MTeX
Here is an example of a script that will load these orientations and plot them in a pole figures. The example is for olivine and can be adjusted to your needs.
- plotGrainPF.m
%% % Define crystal and sample symmetry cs_ol = crystalSymmetry('Pnma',[9.915267 5.886196 4.713400]) ss = specimenSymmetry() % Plotting convention plotx2east % File with Euler angles fname = fullfile('/path/to/folder/',... '','euler.txt'); %% % Load file and create the corresponding list of orientations ori = load(fname) n = size(ori); n = n(1); for i = 1:n phi1 = ori(i,1)*degree; Phi = ori(i,2)*degree; phi2 = ori(i,3)*degree; ori0(i) = orientation('Euler',phi1,Phi,phi2,cs_ol,ss); end %% % Plot grain orientations h = Miller({1,0,0},{0,1,0},{0,0,1},cs_ol) plotPDF(ori0,h,'antipodal','MarkerSize',10, 'MarkerFaceColor', 'b', 'MarkerEdgeColor', 'b')
Loading and plotting grain orientations in MTeX, with colors!
Here is an example of a script that will load these orientations and plot them in a pole figures. The example is for olivine and can be adjusted to your needs.
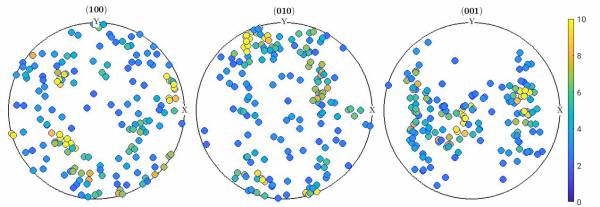
In this case grains are assigned a color based on the ODF intensity for their orientations.
- plotColorGrainPF.m
%% % Define crystal and sample symmetry cs_ol = crystalSymmetry('Pnma',[9.915267 5.886196 4.713400]) ss = specimenSymmetry() % Plotting convention plotx2east % File with Euler angles fname = fullfile('/path/to/folder/',... '','euler.txt'); %% % Load file and create the corresponding list of orientations ori = load(fname) n = size(ori); n = n(1); for i = 1:n phi1 = ori(i,1)*degree; Phi = ori(i,2)*degree; phi2 = ori(i,3)*degree; ori0(i) = orientation('Euler',phi1,Phi,phi2,cs_ol,ss); end %% % Plots grains using a color scale base on ODF fitted to the grains % Fit an ODF to the data odf = calcODF(ori0,15*degree) % Plot orientations, colored based on ODF plotPDF(ori0,eval(odf,ori0),h,'antipodal','MarkerSize',10, 'MarkerEdgeColor', 'b') CLim(gcm,[0,10]) mtexColorbar
Loading and plotting grain orientations in MTeX, in an inverse pole figure, and with colors!
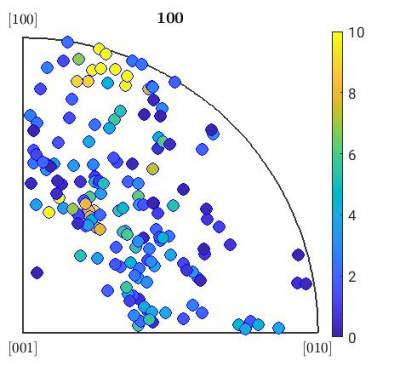
Here is an example of a script that will load these orientations and plot them in a inverse pole figures. The example is for olivine and can be adjusted to your needs. Grains are assigned a color based on the ODF intensity for their orientations.
- plotColorGrainIPF.m
%% % Define crystal and sample symmetry cs_ol = crystalSymmetry('Pnma',[9.915267 5.886196 4.713400]) ss = specimenSymmetry() % Plotting convention plotx2east % File with Euler angles fname = fullfile('/path/to/folder/',... '','euler.txt'); %% % Load file and create the corresponding list of orientations ori = load(fname) n = size(ori); n = n(1); for i = 1:n phi1 = ori(i,1)*degree; Phi = ori(i,2)*degree; phi2 = ori(i,3)*degree; ori0(i) = orientation('Euler',phi1,Phi,phi2,cs_ol,ss); end %% % Plots grains using a color scale base on ODF fitted to the grains % Fit an ODF to the data odf = calcODF(ori0,15*degree) % Plot grains as IPF of the compression (if compression is in X), colored based on ODF plotIPDF(ori0,eval(odf,ori0),[vector3d.X],'antipodal','MarkerSize',10, 'MarkerEdgeColor', 'b') CLim(gcm,[0,10]) mtexColorbar
Comparing grain orientations in MTeX
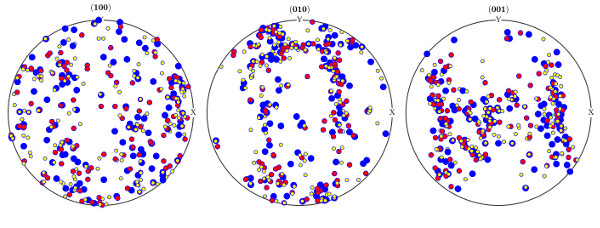
The following script will load results from 3 different indexings a plot the results so they can be compared.
The image below for instance shows the results of a first indexing in blue, a second indexing in red, and a third indexing in yellow.
- compareGrainPF.m
%% % Define crystal and sample symmetry cs_ol = crystalSymmetry('Pnma',[9.915267 5.886196 4.713400]) ss = specimenSymmetry() % Plotting convention plotx2east % Files with Euler angles fname1 = fullfile('/path/to/folder/',... '','euler-1.txt'); fname0 = fullfile('/path/to/folder/',... '','euler-2.txt'); fname2 = fullfile('/path/to/folder/',... '','euler-3.txt'); %% % Load file and create the corresponding 3 lists of orientations ori = load(fname0) n = size(ori); n = n(1); for i = 1:n phi1 = ori(i,1)*degree; Phi = ori(i,2)*degree; phi2 = ori(i,3)*degree; ori0(i) = orientation('Euler',phi1,Phi,phi2,cs_ol,ss); end ori = load(fname1) n = size(ori); n = n(1); for i = 1:n phi1 = ori(i,1)*degree; Phi = ori(i,2)*degree; phi2 = ori(i,3)*degree; ori1(i) = orientation('Euler',phi1,Phi,phi2,cs_ol,ss); end ori = load(fname2) n = size(ori); n = n(1); for i = 1:n phi1 = ori(i,1)*degree; Phi = ori(i,2)*degree; phi2 = ori(i,3)*degree; ori2(i) = orientation('Euler',phi1,Phi,phi2,cs_ol,ss); end %% % Plot grain orientations from the 3 files in a way they can be compared h = Miller({1,0,0},{0,1,0},{0,0,1},cs_ol) plotPDF(ori0,h,'antipodal','MarkerSize',10, 'MarkerFaceColor', 'b', 'MarkerEdgeColor', 'b') hold on plotPDF(ori1,h,'antipodal','MarkerSize',8, 'MarkerFaceColor', 'r', 'MarkerEdgeColor', 'b') hold on plotPDF(ori2,h,'antipodal','MarkerSize',6, 'MarkerFaceColor', 'y', 'MarkerEdgeColor', 'b')